The honey bee is an eusocial species organized in colonies of several thousand individuals. From a genetic point of view, the founding queen of a colony mates beforehand with several males to constitute a pool of spermatozoa which will be used throughout her life to produce the individuals of the colony. Most of the colony is made up of diploid workers resulting from fertilizations and occasionally of a few haploid males, resulting from an unfertilized gamete of the queen and whose role is only reproductive. In beekeeping, traits of interest for selection are mainly measured at the colony level, whereas selection is mainly done at the queen level. It is therefore essential to develop methods that allow to genotype a colony at a lower cost.
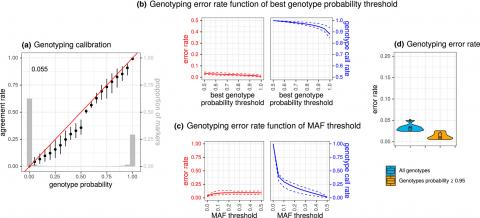
We have developed new statistical models to reconstruct the genotype of the founding queen of the colony from pool sequencing data of a large number of workers. Using data from a reference diversity panel and of the pool-sequencing of a single colony, a first model allows to characterize its genetic composition, i.e. the contributions of the subpopulations to its genetic diversity. From the sequencing of several colonies with the same genetic composition, a second model allows to reconstruct the genotypes of their founding queens. The combination of the two models allows to treat heterogeneous data sets such as those obtained in the research programs of the department's teams. By simulations, we have shown that our models estimate the genetic composition of the colony with an accuracy higher than 90%. The queen genotyping error was estimated at about 2% which corresponds to the genetic information that would be obtained from the individual genotyping of 10 males offspring of the queen. Our approach thus allows to divide by a factor of 10 the genotyping costs of a colony. Among other perspectives, an extension of the models to SNP chip data has been developed.
The model for estimating the genetic ocmposition of a colony in terms of honey bee subspecies uses the supervised version of the STRUCTURE model (Pritchard et al. 2000). The reference data are from the SeqApiPop program (Wragg et al. 2022). The genotype reconstruction model is original and estimates at each marker by maximum likelihood the probability of each of the possible genotypes for the queen of a colony using data available for colonies of similar genetic composition. The models have been implemented in python and are released under an open-source licence (github link).
